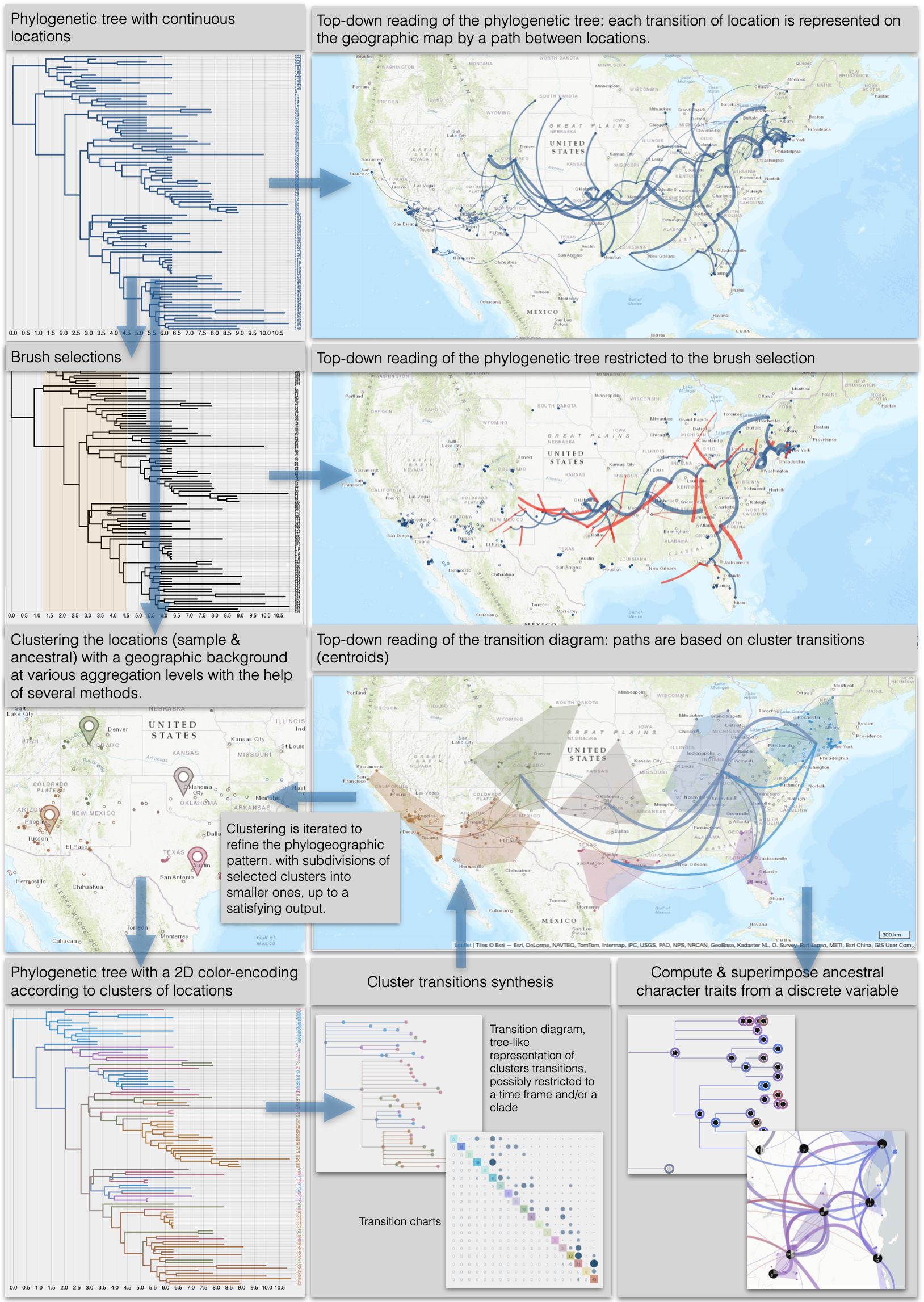
|
EvoLaps aims at visualizing spatio-temporal spread of epidemics, from phylogenetic trees associated with continuous localities (computed ancestral latitude/longitude pairs associated with computed ancestral sequences). Changes of localities ('transitions'), in a "Top-Down" reading of the tree, are represented on a cartographic background using arcs between them. The bundle of arcs is a phylogeographic scenario. EvoLaps helps to analyze complex scenarios:
Questions and comments welcome: francois.chevenet@ird.fr Citation: Chevenet F, Fargette D, Guindon S, Bañuls AL. EvoLaps: a web interface to visualize continuous phylogeographic reconstructions. BMC Bioinformatics. 2021 Sep 27;22(1):463. doi: 10.1186/s12859-021-04386-z. PMID: 34579644; PMCID: PMC8474961. Chevenet et al., 2021 More• User's Manual• FundingThis work has been supported by The PALADIN project, publicly funded through the French National Research Agency under the “Investissements d’avenir” program with the reference ANR-10-LABX-04-01 Labex CEMEB, and support from the Agence Nationale pour la Recherche through the grant GENOSPACE. •Thanksto D. Hubert for system administration and A. Prudhon for English editing |