|
|
||
|
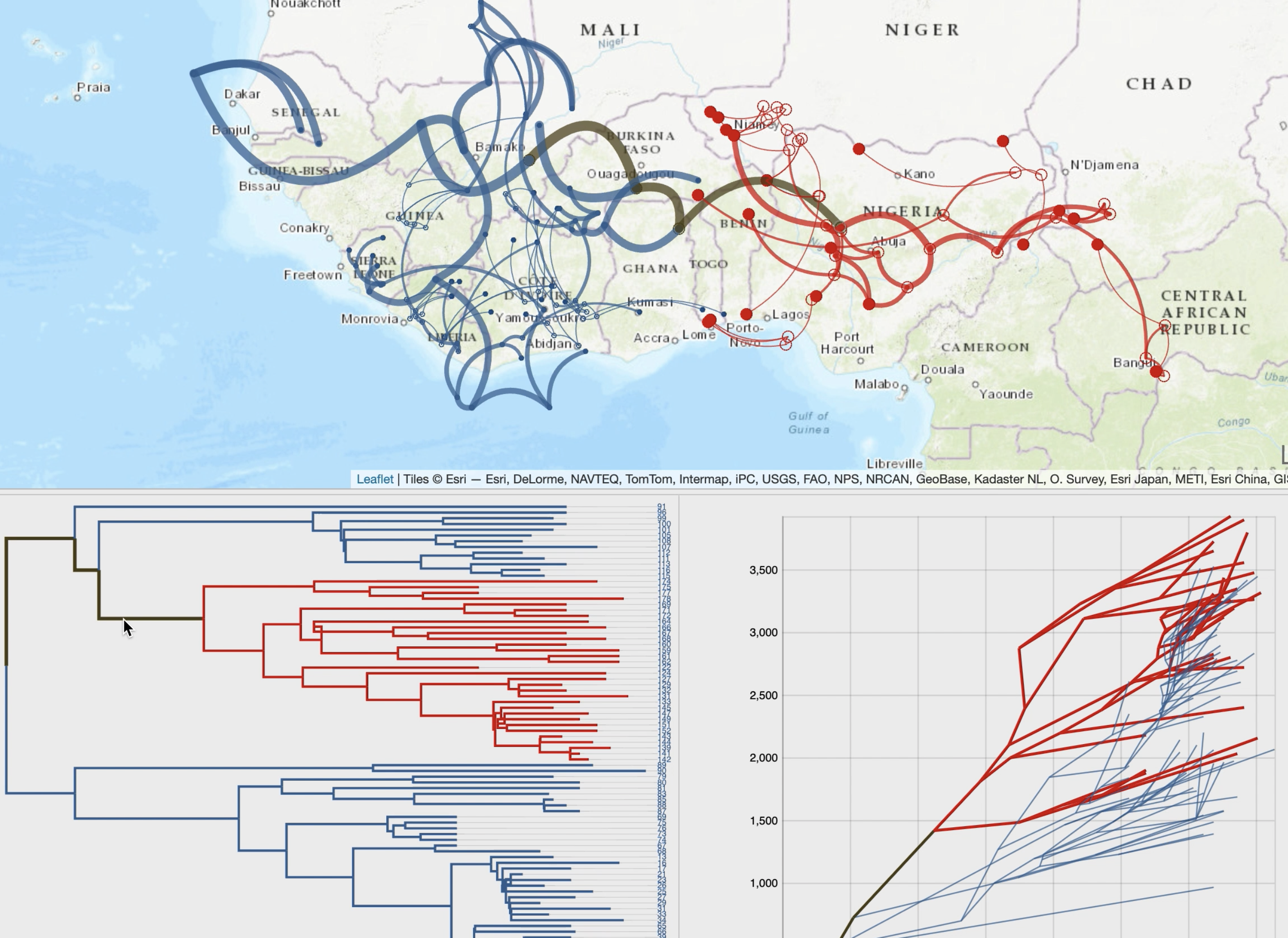
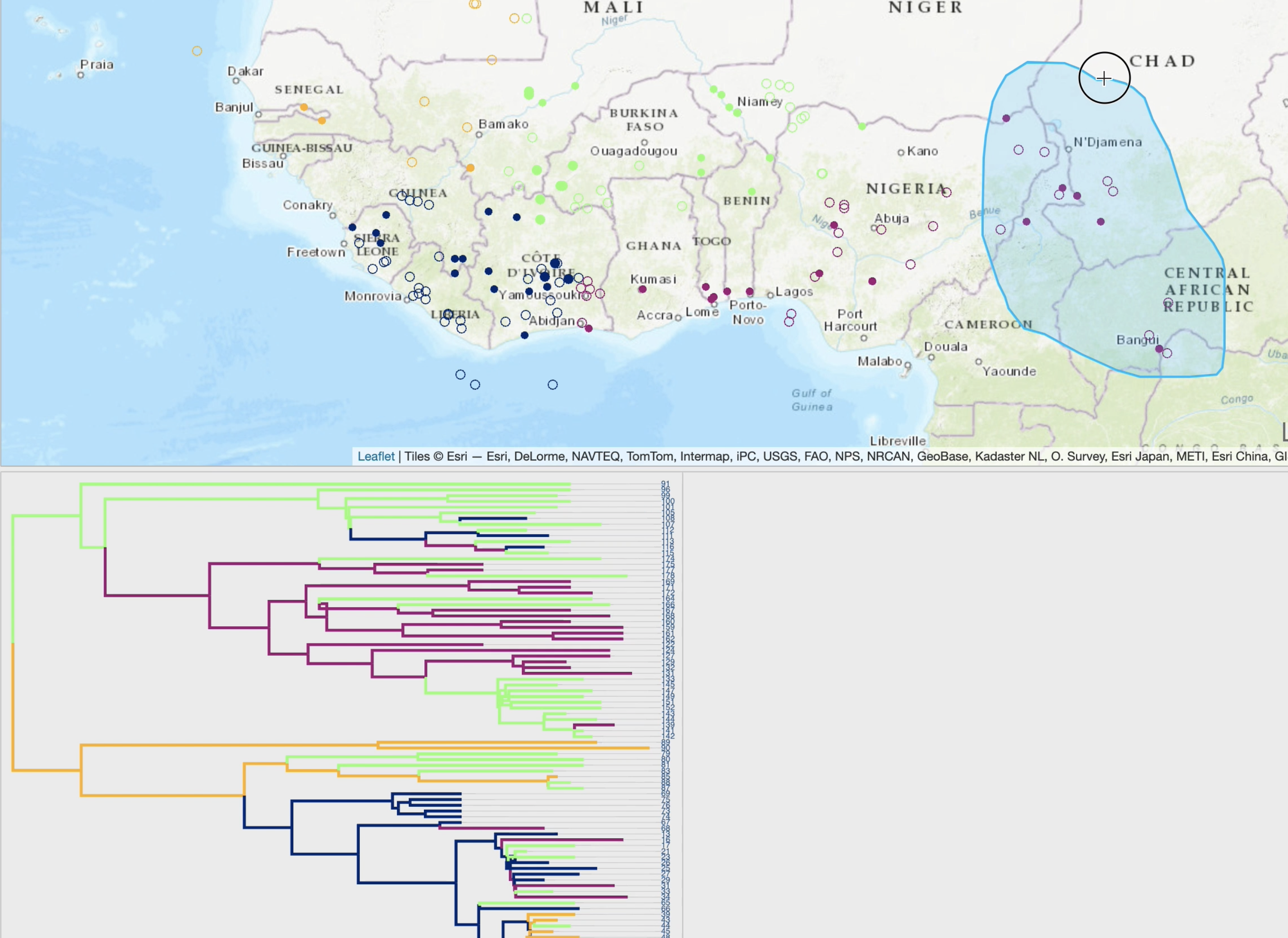
EvoLaps is a user-friendly web application for visualizing spatio-temporal spread of pathogens,
from annotated maximum clade credibility tree obtained through a BEAST continuous phylogeographic inference.
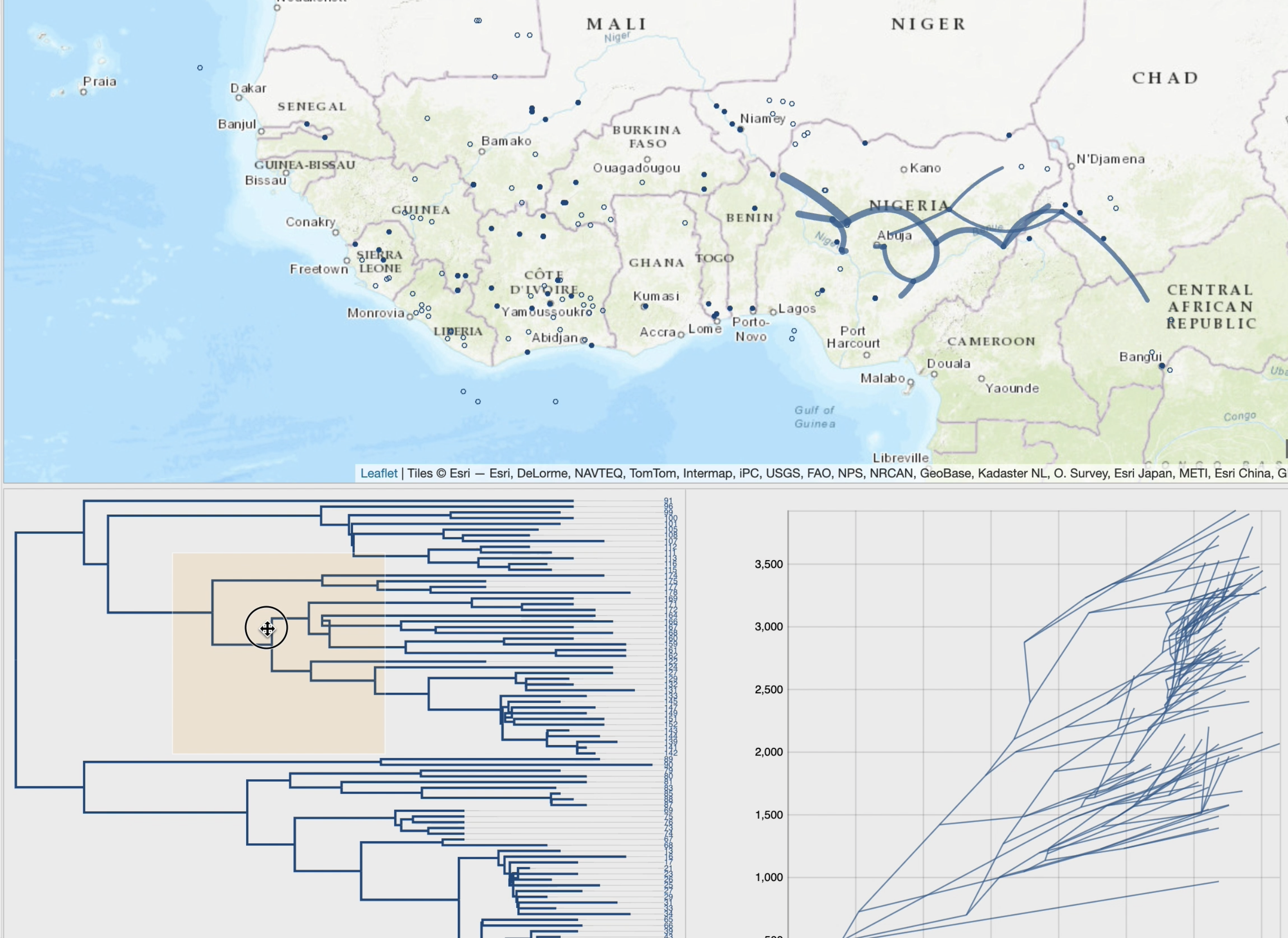
Changes of localities (transitions), in a "Top-Down" reading of the tree (from the root to its leaves in a recursive way),
are represented by paths with a cartographic background. The set of paths is a phylogeographic scenario.
A raw reading of these transitions produces complex scenarios, and EvoLaps helps to analyze them with:
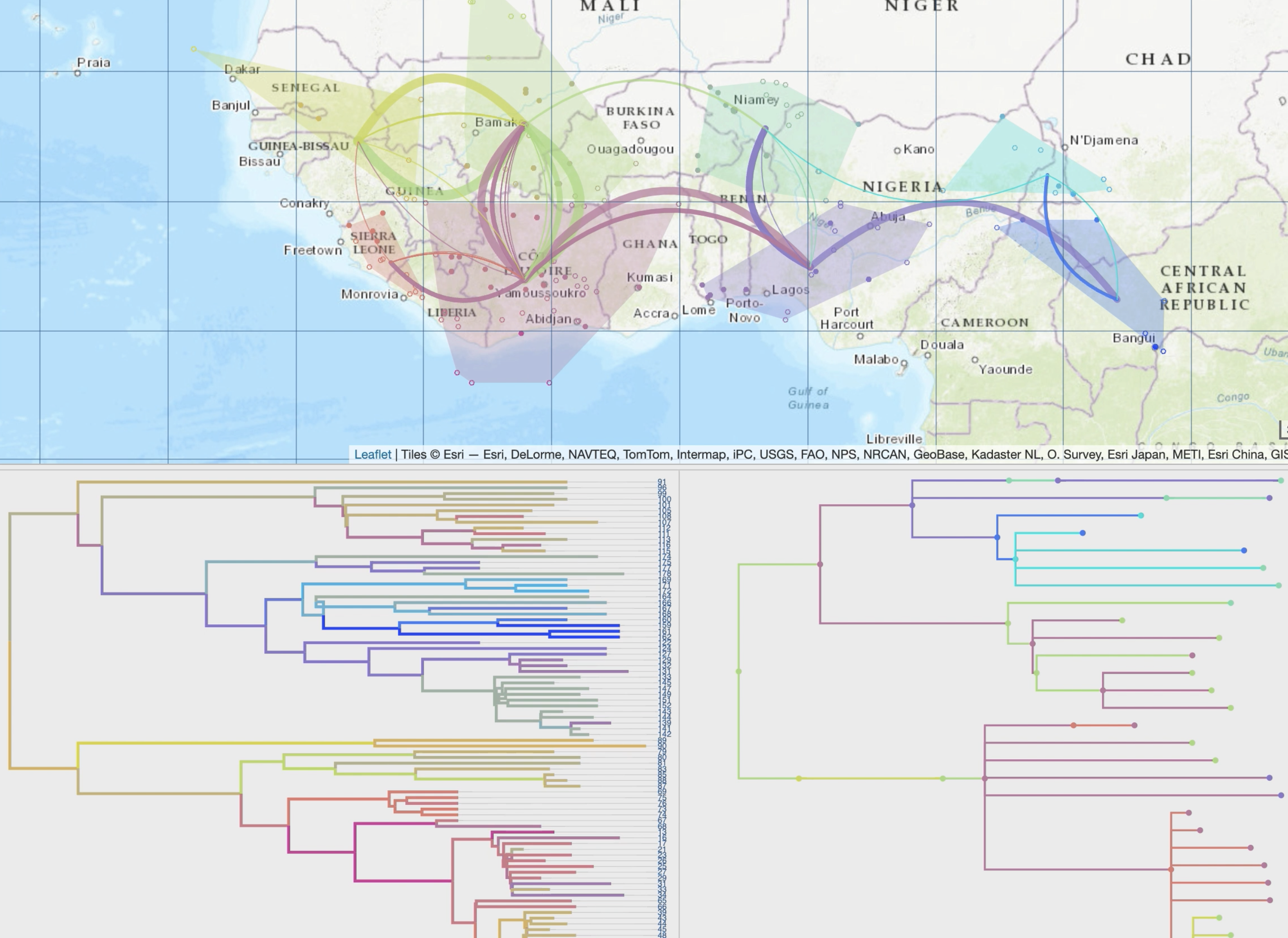
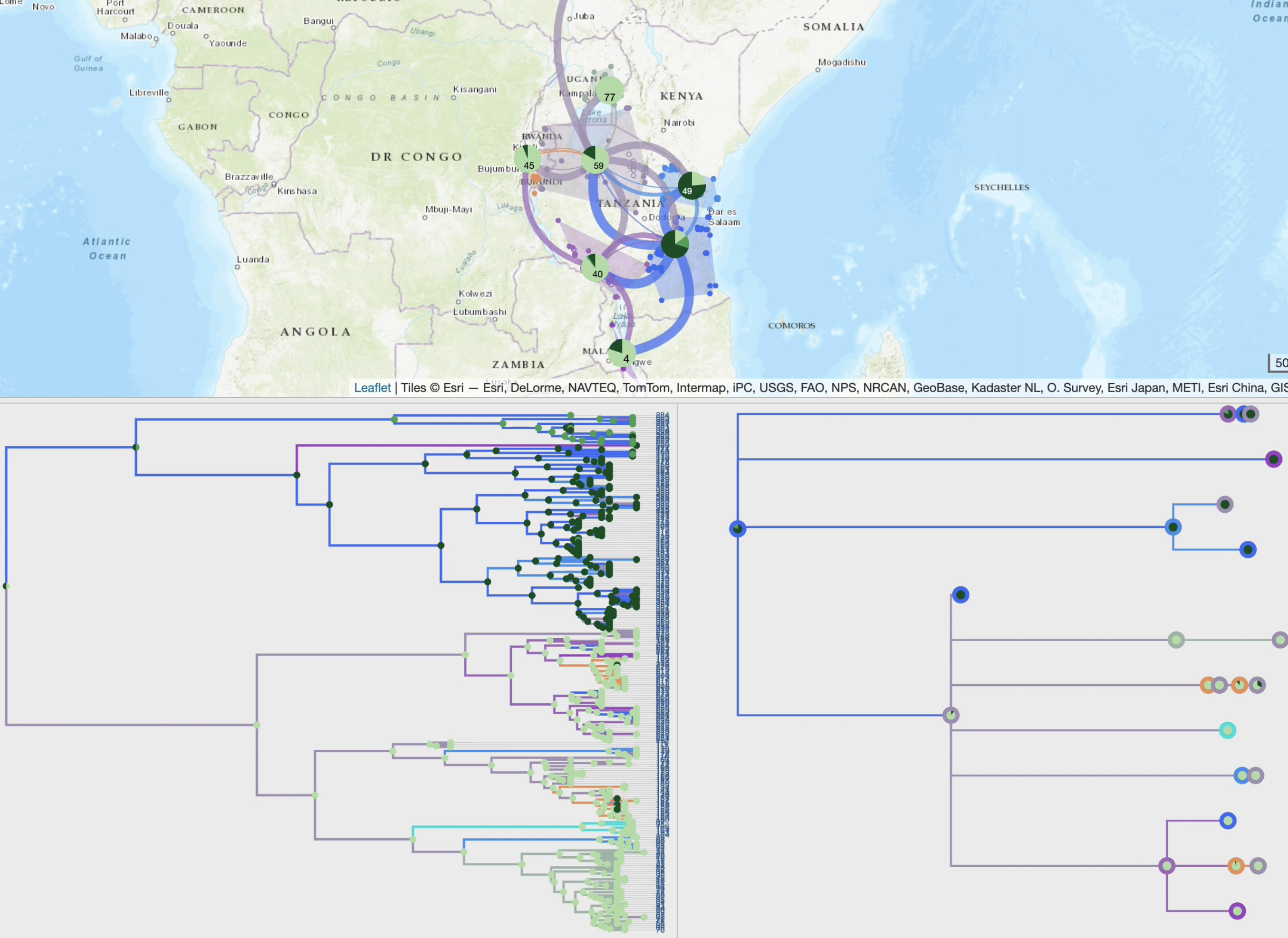
• improved display of paths using multiple graphical variables with optional time-dependent gradients (line thickness, curvature, color, opacity),
• phylogeographic scenario and tree cross-highlight and selection process,
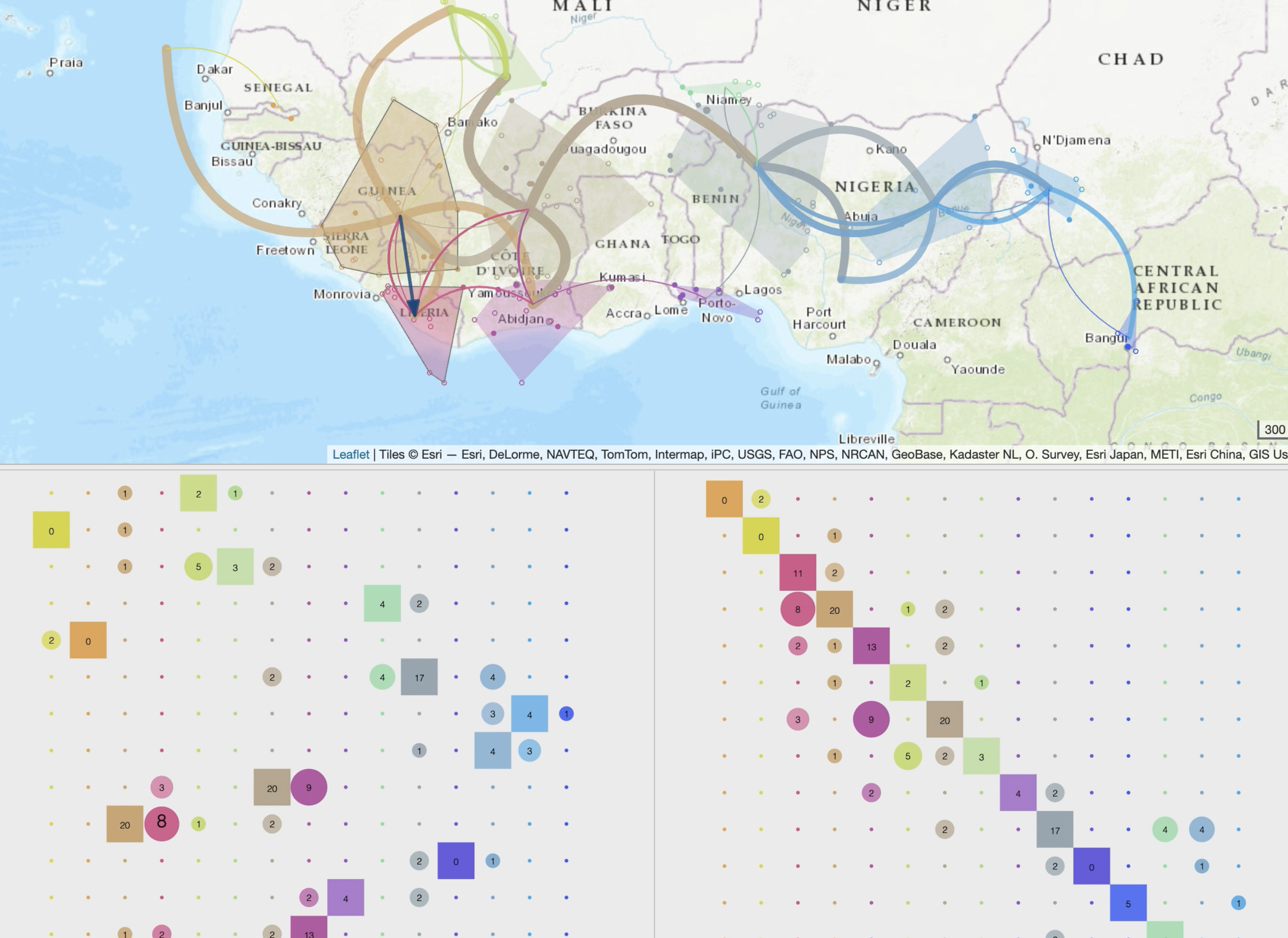
• specific spatio-temporal scales and production of synthetic views based on a dynamic and iterative clustering of localities into spatial clusters,
• phylogeographic scenario animation, based on tree brushing (manual or automatic), gradually over time or at time interval, over the full tree or a specific clade,
• additional views of the phylogeographic scenario through inter-connected modules (migration distance curves, intensity maps, inter-clusters exchange maps or ancestral character states computed from third-party discrete variables).
Contact: francois.chevenet@ird.fr
|
|
|