
EvoLaps
v.39 V40
2
|
+
EvoLaps v.39 Oct. 2023
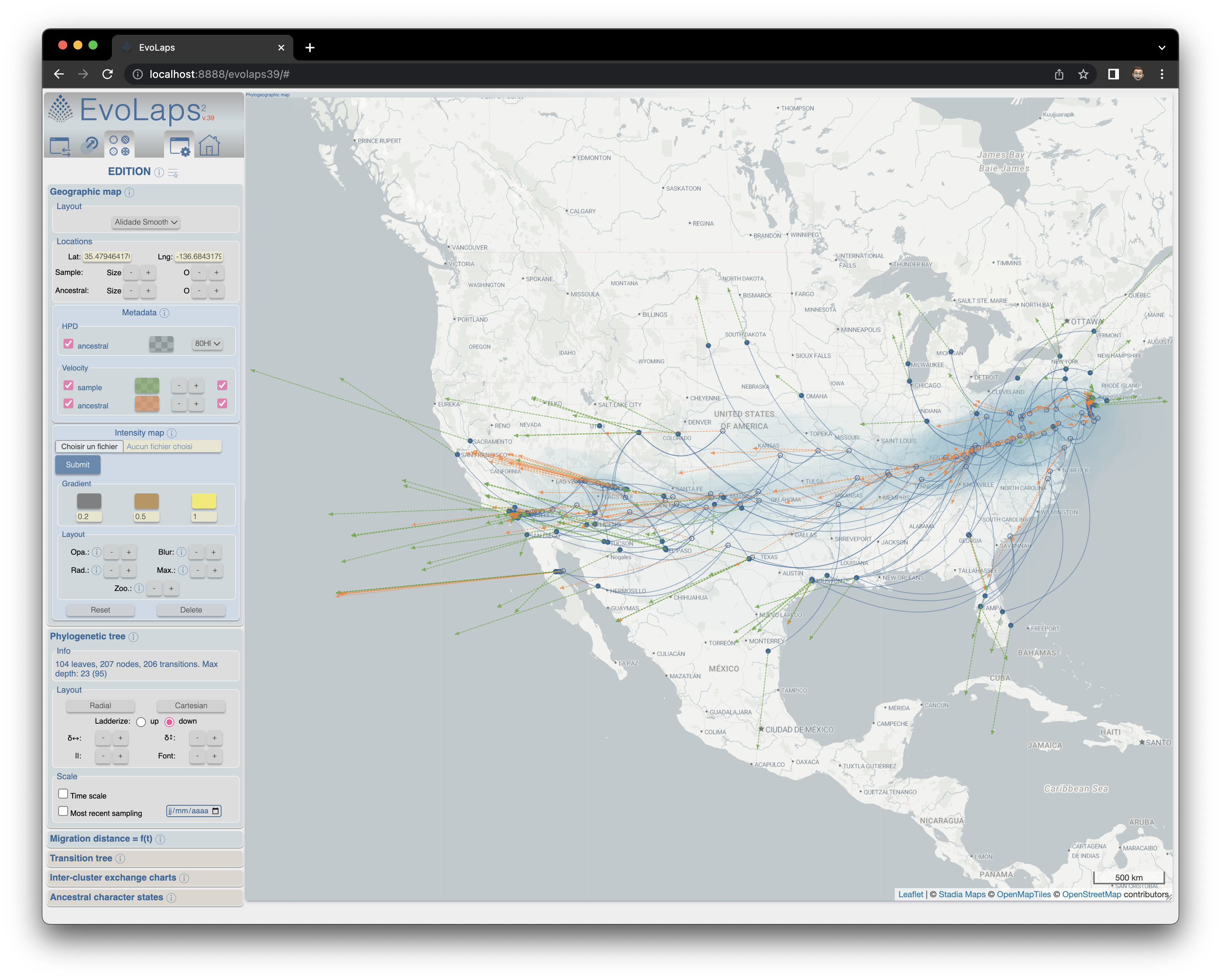
• "Select": new tool enabling the seletion of a node from the phylogenetic tree and display the specific evolutionary scenario for the underlying clade.• New cartographic backgrounds
• Display of Velocity from PhyREX
EvoLaps v.38 Oct. 2023
• compatibilty with the write.beast function from the Treeio package (Wang et al., 2020, MBE)• dynamic HPD treshold menu
Warning
• 'Shift' load EvoLaps to refresh the cache of the navigator. Hold down the 'Shift' key while clicking the reload button, or 'command' key (or CTRL key) + R• Computation of ancestral character sets from discrete variables must be performed using the NEXUS data file format with a clustered analysis as the EvoLaps data file format is currently not functional for this purpose.
Manual
full pdfHow to cite
Chevenet F, Fargette D, Guindon S, Bañuls AL. EvoLaps: a web interface to visualize continuous phylogeographic reconstructions. BMC Bioinformatics. 2021 Sep 27;22(1):463. doi: 10.1186/s12859-021-04386-z. PMID: 34579644; PMCID: PMC8474961. Chevenetet al., 2021API
Icons by Icon8 ⦙ D3 ⦙ Leaflet ⦙ jscolor ⦙ Leaflet-Heat ⦙ Leaflet-Arrowheads ⦙ jQuery ⦙ Bezier.js ⦙ svg-exportJS ⦙ KmeansFunding
• PALADIN project, publicly funded through the French National Research Agency under the “Investissements d’avenir” program with the reference ANR-10-LABX-04-01 Labex CEMEB,• GENOSPACE ANR-16-CE02-0008 from the Agence Nationale pour la Recherche.
Acknowledgments
D. Hubert for system administration and A. Prudhon for English editingPhylogeographic map
Phylogenetic tree

